Almost every day, scientists around the world are discovering new life forms. Archaeologists are digging up past relatives, and evolutionary biologists are adding new branches to the tree of life. While millions of species are estimated to have gone undiscovered to this day, scientists at Yale and Harvard have created a new organism that has wide-ranging applicability in biomanufacturing for industry and medicine alike.
Recoding the Genome
Currently, sequencing the human genome – a process that used to take years – can now be accomplished in ways that are rapid, affordable, and informative. This “decoding” of the DNA blueprint has led to our understanding of specific genes, especially in the context of disease. However, on the opposite end of the spectrum, researchers at Yale, under the direction of Farren Isaacs, Assistant Professor of Molecular, Cellular, and Developmental Biology, has successfully “recoded” the genome of a strain of E. coli< bacteria. Recoding the genome, in its most basic sense, is introducing site-specific changes across the genome in an effort to achieve a phenotypic outcome, such as viral resistance. “We wanted to ask, based on our understanding of the genetic code and certain important properties of the genetic code, such as conservation and degeneracy, could we change it?” said Isaacs. This idea, conceived in 2005 when Isaacs was a postdoctoral researcher in the lab of George Church at Harvard Medical School, led to one of his most recent of his publications in October 2013, entitled “Genomically Recoded Organisms Expand Biological Function” in the journalScience.
The work done by Isaacs and colleagues is predicated on the translation machinery of the cell. As the “central dogma of biology” states, DNA is transcribed into RNA, and RNA is translated into proteins. Isaacs’s research focuses on the latter pathway: translation. During translation, cellular machinery reads the mRNA in three-letter groupings, called codons, in order to correctly add amino acid building blocks to a growing protein chain.
In total, there are 64 codons. Sixty-one of them code for the twenty functional amino acids the cell uses to make proteins; the remaining three codons code for a stop in translation, a signal that the amino acid chain is complete and should be released into the cell to start its life as a protein. These sequences are known as “stop codons,” and are named by their specific base sequences: UAG, UAA, and UGA.
Interestingly, the two proteins responsible for the release of the protein into the body at the end of translation, called “release factors,” have some redundancy in function: they both halt translation upon reaching a UAA stop codon, the most abundant of the stop codons in the E. coli strain that the researchers used in their study. “The codons are discriminated by the tRNA and release factors. Not only do you have to think about redundancy at the DNA level, but also how that gets decoded at the translational level,” said Isaacs.
Two release factors, called RF1 and RF2, operate to release newly-synthesized proteins from the factories manufacturing them. Each one of these factors goes into action in response to two of the three stop sequences that are reached at the end of translation. RF1 is responsible for both UAG and UAA, whereas RF2 is also responsible for UAA and is additionally in charge of the third stop codon sequence, UGA. This means that UAA has the ability to prompt both of the release factors to release the translated protein. It also means that if only two codon stop sequences existed instead of three, a single release factor could be responsible for all of the protein release in the organism.
Isaacs and his team decided to investigate whether one of the stop sequences (UAG) could be recoded – which would involve changing one of the bases in its three-letter sequence – to make it identical to another one of the stop codons, creating an organism with two instead of three stop codon sequences. Decreasing the number of stop codons from three to two would make one release factor extraneous. Isaacs hoped to create a system that simply had no need for a release factor that is undeniably essential to every life form.
CAGE and MAGE: Isaac’s Methods
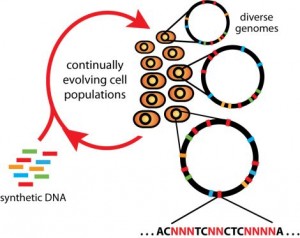
Converting all of one type of stop codon found in the specific strain of E. coli used in the study to a different sequence simultaneously, efficiently, and across the entire genome of a single organism is quite difficult. Isaacs’ new method, known as Multiplex Automated Genome Engineering (MAGE), has allowed his lab to do just that. MAGE employs the use of single-stranded DNA fragments that are ninety nucleotides long, eighty-nine of which are complementary to a sequence found in the genome, and one of which is a “mismatch.”
In the context of this study, the researchers inserted a single nucleotide change that switches the “G” in TAG to an “A” in TAA, effectively converting all TAG’s in the E. coli genome to TAA sequences. In MAGE, a wild-type population of E. coli cells that all have an identical genome is used at the outset. When ninety nucleotide long fragments are introduced, “site-directed” mutations are created in the cell with efficiencies greater than 30%, whereas conventional technologies operate at an efficiency of, at best, less than one percent. These mutations are engineered to occur at a single, specific nucleotide in the DNA of the E. coli genome. Even with such high efficiency, however, the risk of introducing deleterious mutations is quite high. Because a high proportion of E. coli DNA codes for genes, a single undesired point mutation in the genome could result in cell death. Successfully recoding the genome with as few side effects as possible, therefore, requires extremely careful work.
“Just like March Madness, we had 32 initial strains, each of which had a different set of codons recoded,” said Isaacs. Each strain was targeted by Isaacs’ MAGE technology to a different part of the circular E. coli genome, like 32 slices of a pie. “Then the challenge became,” said Isaacs, “‘how were we going to get this into a single organism?’”
That is where a second technology that Isaacs developed, known as Conjugative Assembly Genome Engineering (CAGE), is useful. Isaacs used MAGE to introduce site-directed mutations to convert all TAG sequences to TAA sequences, on 32 separate occasions. CAGE was then used to “merge” the 32 strains, over five consecutive rounds, into a single organism that ultimately contained all 321 TAG mutations. After much work, Isaacs’ team successfully created a genomically recoded organism. In the recoded E. coli they created, the UAG stop codon had been converted into the UAA stop codon, deleting RF1 from the cell, allowing for UAG to lose its ability to “stop” translation. This rendered UAG a “blank codon,” or a kind of canvas for future experimentation.
A Canvas for Innovation
Astonished by the cell’s ability to grow normally under a variety of conditions, Isaacs and his team took the opportunity to use this “blank codon” as an area for expanding the genetic code. While the cell only uses the same twenty standard amino acids, there are several nonstandard amino acids (NSAAs) that can be synthesized, each of which has new chemical properties. Isaacs therefore introduced a new tRNA that is specific for UAG, but loaded with a novel NSAA, in an effort to reassign UAG from a blank codon to a “sense” codon, or one that is capable of coding for an amino acid. Incredibly, the scientists were able not only to recode the E. coli by getting rid of a stop codon that the cell naturally uses during protein synthesis, but they were also able to convert an essential protein, RF1, to a nonessential one, making the UAG stop codon a blank template that they then exploited to incorporate NSAAs.
Throughout this process, the researchers found that not only were these E. coli recoded, but also that, due to the newfound identity of the UAG codon, they became resistant to a bacteriophage, a virus that infects bacteria, known as T7.
“By wiping out the function of an entire codon and because the codon is conserved across nearly every species, the hypothesis was that we could possibly prevent the proper expression of foreign genes that rely on that codon,” Isaacs said. More specifically, the T7 virus was unable to properly translate its infectious proteins, as RF1 was lost in the host cells.
By creating genomically recoded organisms, the scientists successfully produced a “genetic firewall” in which the functional exchange of genetic material between viruses in the environment and the organism is hindered, due to the new translation environment in the host cell.
Future Applications
Although this work has already accomplished significant new feats, many more applications may be possible. In the past, the Isaacs lab has applied MAGE commercially to mass-produce an anti-cancer antioxidant at an unprecedented speed in E. coli. When he was asked, Isaacs affirmed the fact that there is much industrial interest in his work and in the innovative methodologies he has invented. With the introduction of synthetic amino acids, these genetically recoded E. coli have the potential to produce new biomaterials that have uses for entities like drug delivery vehicles and the U.S. army.
The possibilities this research opens up are tremendous. While some believe that this is the start of a Frankenstein-esque, sci-fi future of genetically recoded organisms, others, like Isaacs himself, are enthusiastically optimistic. “Biology is the most powerful technology we have,” says Isaacs. Biology, unlike the hardwired structures of computers, is a dynamic, evolving system that is only beginning to reveal its vast potential as a tool for significant engineering innovation.
Cover Image: Art by Nicole Tsai

