Since identifying HIV in the mid-1980s, scientists have been trying to better understand the enigmatic qualities of this virus. Current research is approaching the issue by trying to more precisely characterize the relationship between the virus and the body. Researchers in Yale’s Molecular Biophysics & Biochemistry Department have revealed the crystal structure of the myxovirus resistance protein B, which has been shown to restrict HIV-1 (the form of HIV that causes AIDS). This finding by Professor Yong Xiong and his team helps explain why and how this specific protein inhibits the HIV virus whereas other similar proteins do not.
HIV attacks a person’s immune system, specifically their CD4 T cells, reducing one’s ability to fight infection and disease. If HIV destroys too many of these cells, patients eventually develop AIDS. Though some treatments exist, no cure has yet been discovered for either the virus or the syndrome. Over 33.4 million people in the world currently live with HIV/AIDS, and there have been over 25 million reported deaths from AIDS since its documentation. Because of this, it is of particular interest to the scientific and medical communities to better understand the mechanisms behind HIV virus infection and how to target it.
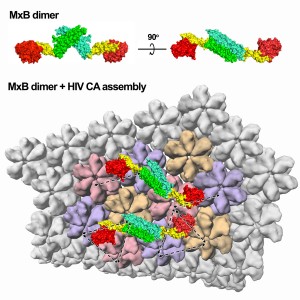
HIV is composed of only 15 proteins and two strands of RNA inside its capsid (protein shell). Humans have two myxovirus resistance (Mx) proteins, which are able to inhibit various viruses including HIV. MxA and MxB are 63 percent identical, but function in very different ways. Previously, it was demonstrated that MxB is able to hinder HIV-1, but that MxA did not have this capability. Why this difference exists between the structurally comparable proteins was unknown, motivating Xiong’s lab to explore the causes. By deciphering the crystal structure of MxB using X-ray crystallography, researchers were able to more clearly deduce the determinants involved in HIV-restriction.
Because MxA and MxB are so alike, any differences between them may signal key structural differences that are responsible for their disparate functions. MxB was found to be able to function as a dimer (two MxB linked together) whereas MxA needs to be in a large ring-like form to operate. Another difference discovered between MxA and MxB is that MxB does not need to get energy by breaking down a molecule known as GTP to inhibit HIV, whereas MxA’s utility does depend on this enzymatic activity. The inherent architectural difference between the two proteins in their active form explains their varied mechanisms. Additionally, it was determined that MxB can bind directly to the HIV capsid, which could possibly block uncoating of the virus or prevent nuclear import of the viral genetic material.
Analyzing the mechanisms involved in protein inhibition of the virus can help lead to better HIV treatment development in the future. “The idea is to better understand the virus so that we can design something to counteract the viral infection,” explained Yong. “We already have an anti-viral agent in our body, we just need to learn how to harness its power to increase its potency against the virus in order to have a natural solution.” Alternatively, Xiong added, another option would be to “design a drug that acts in a similar way to MxB to inhibit the virus.” The better scientists are able to comprehend how the virus interacts with the host, the better they will be able to target it to prevent infection.
