
In 1975, Frederick Sanger developed a sequencing technique using DNA polymerase to radioactively label nucleotides. This procedure was called the “Plus and Minus” technique. Forty years later, full-genome sequencing lies at our fingertips. We have sequenced the genomes of many important model organisms, including the fruit fly (Drosophila), roundworm (C. elegans), and house mouse (Mus musculus). We even finished sequencing the human genome in 2003.This year, another organism joined the honored ranks, and it was…the tsetse fly?
Considering the damage it does, not many people know about the tsetse fly. The insect resides in sub-Saharan Africa, and it is the primary transmitter of a parasite called trypanosome, which causes sleeping sickness. The fly feeds on blood, so it can transmit the parasite directly into the bloodstream of its victims. Symptoms of the disease include fever, anemia, lethargy, and eventually, death. The fly infects approximately 20,000 people a year and puts another 70 million people at risk. Sleeping sicknessalso affects livestock, so the ongoing epidemic has hindered the sub-Saharan African region economically. It is no wonder that scientists are looking for ways to limit the devastation caused by the tsetse fly.

As surprising as it was, the full-genome sequencing of the fly has already inspired numerous solutions to reduce the incidence of sleeping sickness in sub-Saharan Africa. Previous methods to control the tsetse population ranged from burning its habitats to manipulating its sex cycle by breeding sterile male flies and having them mate with female flies. While these techniques have helped, the sleeping sickness epidemic has continued. Now, scientists have jittery hands to try out the new gun in the holster: the tsetse fly genome. With this tool, scientists have concocted solutions that exploit the reproduction, digestion, and vision of the tsetse fly.

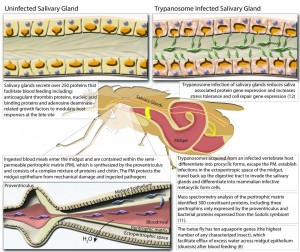
Dr. Serap Aksoy, Professor of Epidemiology at the Yale School of Public Health, spearheaded the tsetse fly genome project, which involved 146 scientists from around the world. Her findings were published in the April issue of Science. In the study, the team discovered that the tsetse genome consists of 366 million base pairs, one-tenth of the human genome, which has approximately three billion base pairs. More importantly, of the fly’s thousands of protein-encoding genes, 12 genes code for proteins involved in milk production. Female tsetse flies depend on milk to nourish progeny in the womb. Researchers hope to develop a drug interrupting these proteins, which would stifle the fly’s reproductive cycle.
Furthermore, the team noticed that the tsetse fly lacks genes coding for gustatory receptors associated with sweet taste. Most flies have these receptors, but the tsetse relies solely on blood rather than any other plants to obtain nutrients. Aksoy believes that blood can be a mode of distributing tsetse fly-specific insecticide. If livestock are vaccinated so they produce antibodies to combat digestive proteins found in the fly’s gut, the fly will receive only limited nutrients when it bites a cow. Malnutrition will then prevent the fly from breeding.
A final genetic discovery that scientists hope to exploit is the presence of the Rhodopsin 5 (Rh5) gene. This gene expresses high amounts of blue-sensitive photoreceptors, and thus explains the fly’s attraction to the color blue. Scientists will be able to use this fact to develop more efficient lures that are fine-tuned to the sensitivity of this photoreceptor. As such, tsetse flies will be more efficiently captured and contained.
This publication on the genome of the tsetse fly demonstrates the importance of genomics research in targeting health issues. In one fell swoop, Aksoy and her colleagues were able to uncover multiple points that could be utilized to control tsetse populations. With more genetic information at the hands of innovators, it is more likely that scientists will tackle lesser-known, neglected diseases such as sleeping sickness. Because the disease occurs primarily in sub-Saharan Africa and is overshadowed by regional health problems such as HIV/AIDS and malaria, preventative measures for sleeping sickness are scarce and diagnosis is complicated. If patients with sleeping sickness are not properly treated, there is a 100 percent mortality rate.
Genomic sequencing of the tsetse fly provides scientists the opportunity to fight back against this deadly disease. More generally, genome sequencing has shifted slower hypothesis-based research to information-based research, and has offered a new wealth of information to scientists. In this day and age, science must turn towards innovation to confront pressing global health problems. We see how quickly ideas and solutions to contain tsetse fly populations have sprung from knowing the organism’s genome. Who knows what else we can discover from genome sequencing?
