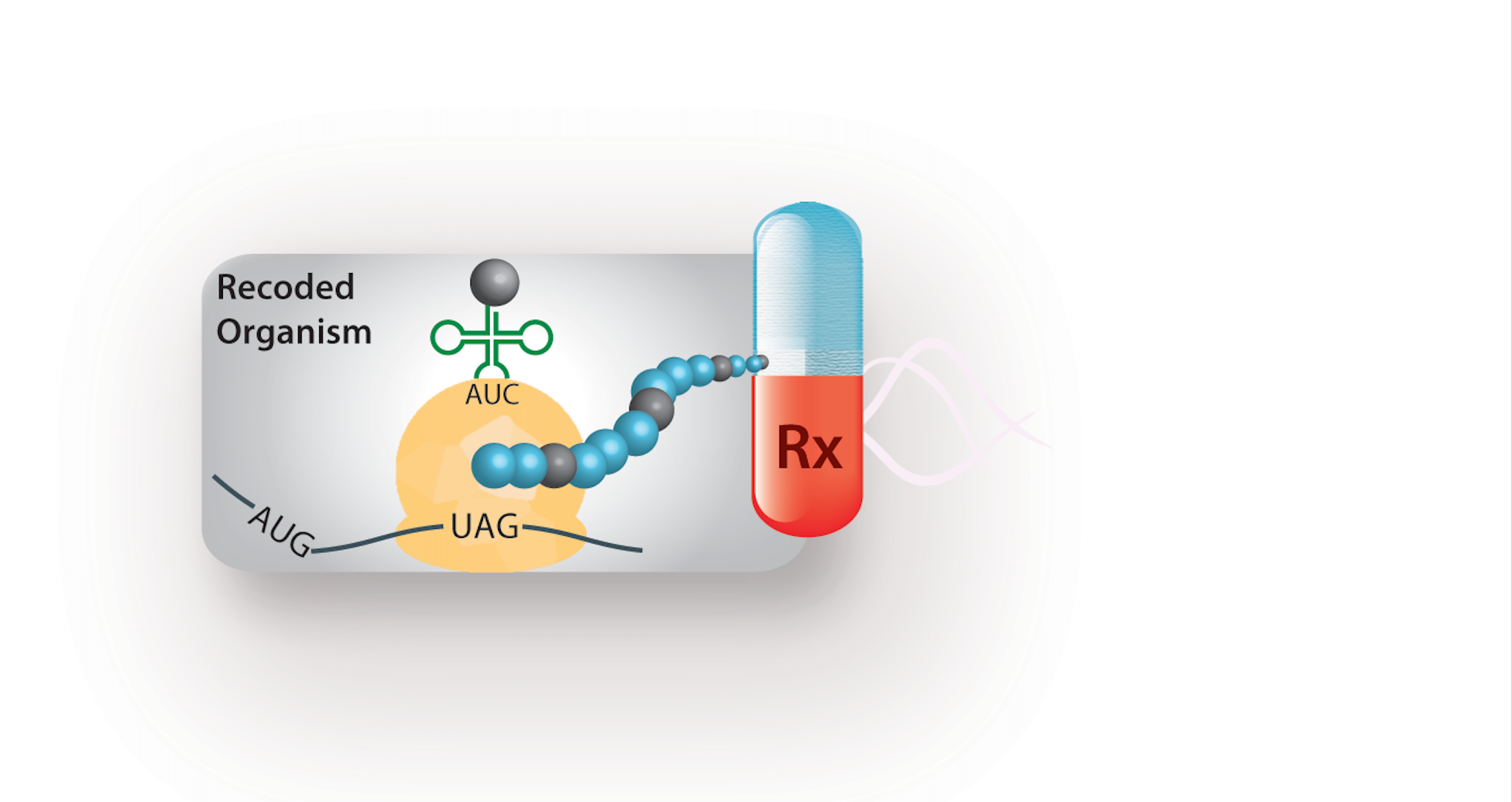
Bacteria have long been utilized as molecular factories. Scientists can tinker with the DNA of these organisms such that they produce proteins of medical or industrial significance in large quantities. However, researchers have been limited to using protein building blocks—amino acids—that naturally occur in living things. Many laboratories have attempted to reprogram these ‘factories,’ creating structures that consist of units other than the twenty naturally occurring amino acids. For the past twenty years, these labs have been limited to just a few non-standard amino acids per protein. A team led by Farren Isaacs, assistant professor of molecular, cellular, and developmental biology at Yale University, managed to improve upon these techniques, allowing for non-standard amino acid incorporation at multiple sites in a protein.
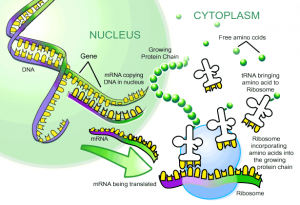
To create these more advanced bacterial ‘factories,’ Isaacs and his team used the fact that the genetic code is degenerate, with multiple ‘words’ coding for the same building block. The information carried in this code is transcribed into messenger RNA (mRNA) molecules, which in turn provide instructions to ribosomes, the sites of protein synthesis. Three consecutive mRNA bases—known collectively as a codon—code for a single amino acid. Because there are more codons than there are amino acids, this means that scientists can modify the genetic code by taking one of the redundant codons and using it to encode a non-standard, or synthetic, amino acid. Though this process sounds relatively simple, there were many fine points that the researchers had to iron out.
To actually build a protein on the ribosome, the specific codons on mRNA molecules each signal for the addition of specific amino acids. These incoming amino acids are each attached to unique transfer RNAs (tRNAs) that latch onto the ribosome and help to stitch together the nascent polypeptide. Each of the amino acids is attached to its correct tRNA via an enzyme known as amino-acyl tRNA synthetase. In order to synthesize proteins with synthetic amino acids, the team needed to repurpose this enzyme, such that it efficiently attached a synthetic amino acid to a corresponding tRNA. Until now, this has been extremely difficult to do in the lab.
Isaacs and his team solved this problem by drawing inspiration from evolution. Over long periods of time, as organisms mate and reproduce, random changes (or mutations) occur in the genetic code. Over time, nature selects for organisms with beneficial mutations, but selects against those with deleterious ones. “We drove the molecular evolution of translational components to enhance their performance,” Isaacs said. The team evolved the amino-acyl tRNA synthetase enzyme: they mutated the E. coli bacteria to create a variety of mutant forms of this enzyme. They then selected for organisms in the pool of mutants that were better equipped to attach synthetic amino acids to their tRNAs.
Because of the increased efficiency of their evolved enzyme, Isaacs and his team were able to induce bacteria to incorporate multiple synthetic amino acids into proteins with high yield and fidelity. The applications for this work are remarkably far-reaching.
“Previous to [this project], the state-of-the-art technology allowed us to integrate one to a handful of [synthetic] amino acids at a time, and that is at low yield and low purity,” Isaacs said. “We are now able to produce entirely new types of sequence-defined synthetic polymers.”
One such type of polymer is a carrier for protein-based drugs. These drugs are very specific and potent, but are extremely small in size. As a result, they are sometimes degraded and cleared too rapidly, before they can be fully efficacious. Isaacs and his team hope that multi-site incorporation of synthetic amino acids will help to more creatively manipulate the structures of these drugs to address this issue.
The expression of proteins with nonstandard amino acids may also be relevant in the field of material chemistry. The formation of aggregates of such proteins could provide better nontoxic adhesives for use in surgery or dentistry.
Though the applications of this creative engineering strategy are still subject to speculation, the possibilities are endless. There is no doubt that it will help to improve the lives of many.