Discovering the evolvability of RNA structures
We often think of “evolution” as the work of millennia: a slow but steady process by which species diversify and gradually adapt to different environmental niches. On a much smaller scale, evolution must also result in biochemical machines that fine-tune an individual’s response to its environment—machines that can themselves be easily adaptable. A team of Yale researchers, led by Professor Scott Strobel of the Yale Chemical Biology Institute, recently identified how RNA can be key to this fast and precise recoding of these machines.
Just like a grocery list helps you buy exactly the ingredients you need only for the meals you want to make, cells use a “grocery list” of chemical signals to regulate gene expression and use their energy most efficiently. In fact, an mRNA molecule, which acts as an intermediate between genes and protein synthesis, can interpret these signals to control its own synthesis using structures called riboswitches. Each riboswitch includes an aptamer (binding site) that binds one specific chemical, or ligand. The ligand is closely related to the functions of whatever protein that mRNA creates, and its binding turns mRNA creation on or off. But how were these precise chemical input-output relationships coded?
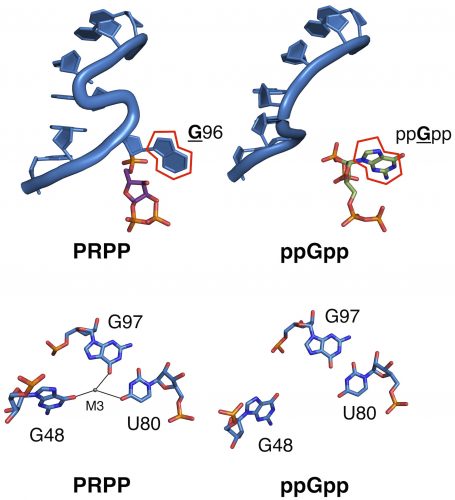
Researchers in the Strobel Lab have identified a set of bacterial riboswitches that can evolve easily, showing functional “ruggedness.” In their study, switching just a single nucleotide, the fundamental unit of RNA, allowed two otherwise identical aptamers to bind to dramatically different ligands with high specificity.
The researchers used X-ray crystallography to characterize two riboswitch-ligand complexes—a painstaking technique due to the difficulty of crystallizing biomolecules. The researchers must guess the conditions in which the molecule is neither dissolved nor fully solidified by doing a “sparse matrix screen”—what Strobel lab member Andrew Knappenberger described as “a giant n-dimensional game of Battleship.” It’s as hard as it sounds.
However, the struggle to crystallize changed the course of this study. Knappenberger and Caroline Reiss, another member of the lab, had been independently trying to crystallize two related riboswitches, which shared a common sequence that differed only by one nucleotide. When Knappenberger got a “hit” on his sparse matrix screen, Reiss decided to try to crystallize her ligand with his riboswitch, mutated by that single nucleotide. Incredibly, it worked.
In both riboswitches, the researchers found, the ligand binds at a structure called an S-turn. Reiss’s single nucleotide mutation occurred in this specific region, causing a change in what ligands that area could now bind. They then quantified the change in binding affinity for the new ligand and found an estimated affinity switch of 40,000-fold.
This research shows that understanding the structure and function relationships of riboswitches can provide a highly responsive tool for synthetic biology: researchers can use riboswitches as to easily control the expression of different genes.
This new evidence of RNA adaptability may also shed light on evolutionary history. Recoding the function of a riboswitch requires a change in ligand specificity that is somehow evolutionarily advantageous. The results of this study demonstrate that acquiring this new specificity is possible with just a single nucleotide mutation. Other conserved RNA structures may be similarly “rugged”— a tantalizing hint at the role of RNA evolution in the origin of life.