The term ribonucleic acid – RNA – conjures up images of a nucleic acid that merely serves as a mediator of information between DNA and proteins. It is seen as just a second step in the pathway to protein synthesis, and because of this dismissal of its significance, RNA research for decades was not assigned the same importance as that of DNA or proteins.
However, a few major breakthroughs led to a radical shift in the perception of RNA’s role in the body. The discovery of ribozymes by Yale professors Thomas Cech and Sidney Altman showed that RNA could catalyze reactions. This led to the proposal of the RNA world hypothesis, which claimed that RNA was the original nucleic acid because it contained catalytic properties, and that DNA eventually evolved from RNA.
In the past decade, the unearthing of small RNAs led to an explosion in the fields of biomedical research and medicine, and was dubbed the “Breakthrough of the Year” by Science magazine in 2002. The existence of small RNAs revealed that RNA plays a much more active role in regulating cellular processes than once assumed.
Dr. Haifan Lin, Professor of Cell Biology and Director of the Yale Stem Cell Center, capitalized on this newfound role of RNA in the body. Together, Lin and his graduate student, Hang Yin, discovered piRNAs, a subclass of small RNAs that play an integral role in regulating gene expression.
What is RNA?
RNA is a single-stranded nucleotide chain comprised of four nucleotides: adenine (A), guanine (G), cytosine (C), and uracil (U). It is synthesized by RNA polymerase using a DNA strand as a template during transcription, matching A with U, G with C, and vice versa.
Once the RNA has been synthesized, it can be grouped into one of several categories. Messenger RNA (mRNA) serves as an intermediary from DNA to protein. This type of RNA will ultimately be transported to the cytoplasm and translated by ribosomes into proteins.
Another major class includes functional RNAs, which directly play roles in diverse cellular processes. For example, transfer RNA (tRNA) molecules transport amino acids to the ribosome during translation. Ribosomal RNA (rRNA) forms the major structural components of ribosomes.
The recent discovery of a group of functional RNAs, small RNAs, has single-handedly altered the discussion surrounding the central dogma of biology. This tenet holds that DNA codes for the transcription of RNA, which then is translated into a protein; yet small RNAs are capable of halting the translation of RNA, or even the transcription of DNA.
How do small RNAs regulate large DNA molecules?
Small RNAs are just 21-28 nucleotides long, but they are responsible for regulating the expression of particular genes. Yin explained that the discovery of small RNAs “initiated a whole new field of non-coding RNA studies, in that these non-coding small RNAs have important roles in controlling gene expression and preventing host genome from foreign DNA invasion.”
In 1998, Andrew Fire and Craig Mello published a paper in Nature describing a gene silencing effect after injecting double stranded RNA (dsRNA) into the roundworm C. elegans. They noticed that the dsRNA successfully attenuated production of a muscle protein, thereby silencing the targeted gene.
The mechanism for this silencing is known as RNA interference (RNAi). An enzyme called Dicer cleaves dsRNA into smaller RNA fragments, now called small interfering RNAs (siRNAs). The siRNAs enter the RISC complex, where they then base-pair with complementary target mRNA molecules, leading to the enzymatic degradation of the mRNA transcript.
Scientists have shown that RNAi evolved as a mechanism to resist viral infection in the plant genome. Another class of small RNAs, micro RNAs (miRNAs) are related to siRNAs, except they are processed from single-stranded precursor molecules and show only partial base complementarity to their targets. The piRNAs discovered in Dr. Lin’s lab play a role in gene silencing in mammalian cells.
What are piRNAs and how do they control gene expression?
PiRNAs are longer than average miRNAs that are expressed in the spermatogenic cells in the testes of mammals. They originate from a region of highly packed chromatin called heterochromatin, characterized by its silenced state of transcription and its highly repetitive sequences.
The active state of chromatin, euchromatin, is characterized by lightly packed DNA often undergoing transcription. Lin investigated the role of piRNA in epigenetic regulation – the switching between the heterochromatic and euchromatic states.
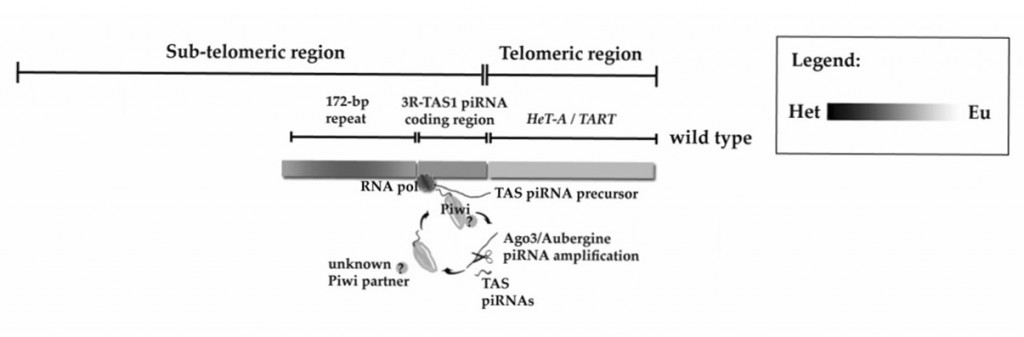
Central to his research is the PIWI protein, which plays a role in RNAi, the maintenance of germ-line stem cells, heterochromatin formation, and transposon silencing. Lin studied the effect of piRNA and PIWI protein on the telomere-associated sequence (TAS) on the right arm of chromosome 3 (3R-TAS) using the fruit fly, Drosophila melanogaster, as the model organism.
In order to identify piRNAs that interact with PIWI, 19,048 candidate small RNA clones were obtained. At first, Yin purified and sequenced piRNAs using a method called Ligation-mediated RT-PCR, a standard method for characterizing miRNAs.
However, he soon realized that while this method was suitable for miRNAs, since there are only several hundred in the genome, it proved vastly inefficient for piRNAs, of which there are an estimated 200,000! Lin then switched to high-throughput sequencing, which allowed sequencing of millions of short sequences simultaneously, and allowed him to identify 12,903 PIWI-associated piRNAs.
Next, it was crucial to verify the direct binding of PIWI to piRNAs in vitro. This binding showed that the PIWI-piRNA complex could indeed exist and perform a vital epigenetic function in vivo.
Third, the overall epigenetic effect of a PIWI mutation in the genome was evaluated. In PIWI mutant flies, levels of common heterochromatic modification markers were considerably increased, whereas those of common euchromatic modification markers were decreased. Taken together, these results suggest that PIWI, along with piRNA, promotes euchromatin formation and heterochromatin silencing.
Finally, Lin and Yin set out to create a cohesive model to explain the effect of PIWI along with the 3R-TAS piRNA, leading them to postulate a heterochromatin/euchromatin counter-balance model. In their model, the TAS region of chromosome 3 transcribes a piRNA precursor in wild type flies, which is then processed into the actual 3R-TAS piRNA, and finally loaded onto a PIWI complex, which is guided to 3R-TAS. An unknown component in the complex then prevents heterochromatin formation and promotes transcription of the piRNA precursor.
“This is a very specific model,” Yin explained, “and we have no idea whether the model might be applied to other genomic regions or other model organisms.” On a larger scale, they postulate that the PIWI-mediated piRNA pathway might regulate global chromatin organization.
An intriguing consequence of this research is that it makes us reevaluate what is traditionally considered “junk DNA.” Yin explained that 60% of piRNAs are derived from transposable elements, which are usually deemed as “parasitic foreign DNAs or junk DNAs” in our genome.
Transposons are sequences of DNA that can move around to different locations within the genome in a cell. Because of this property, they are highly capable of mutagenic activity, since they can insert themselves within a gene, causing dysfunction, or create a gap in the genome after leaving, which is not properly repaired.
The 3R-TAS region regulated by piRNAs bears sequence homology with ancient transposable elements. PIWI and piRNAs are necessary for the proper epigenetic regulation of 3R-TAS, and in turn, the maintenance of germline stem cells. Therefore, this research raises the question of whether our genome has assimilated the transposable elements and harnesses them for important biological functions.
The future of piRNAs
Additional findings point to piRNAs exerting opposite roles in different parts of the genome. Joan Steitz, Sterling Professor of Molecular Biophysics and Biochemistry, recently discovered that miRNAs can upregulate translation, a divergence from their typical role of repression.
As Yin explained, “Our findings start to reveal the complexity of small RNA-mediated epigenetic regulation. Namely, PIWI protein can exert opposite effects on different genomic regions.” He plans to systematically investigate where PIWI exerts its activation and silencing functions in the genome, and also look into what causes the “bipolar” nature of PIWI.
In the budding field of piRNA research, Lin and Yin’s work is a stepping stone to helping answer many questions regarding the epigenetic regulation of the genome.
About the Author
Vikram Jairam is a sophomore in Morse College majoring in Biology, and is excited about the prospects of piRNA in biomedical research.
Acknowledgements
The author thanks Dr. Hang Yin for taking the time to thoroughly answer his many questions on the subject of piRNA.
References
Lin, Haifan., Yin, Hang. An epigenetic activation role of PIWI and a PIWI-associated piRNA in Drosophila melanogaster. Nature 450, 304-308 (2007)
Couzin, Jennifer. Breakthrough of the Year: Small RNAs. Science 298, 2296-2297 (2002)