
Deep inside a computer, past the screen and keyboard, past the motherboard and circuits, beyond the operating system and software, are the zeroes and ones of binary information storage. Likewise, biological systems such as DNA can be deconstructed. This extraordinary molecule encodes the information for life in a system of four nucleotide bases: A, T, C, and G. But the distinction between genetic and machine code is fading. Nick Goldman and Ewan Birney of the European Bioinformatics Institute have managed to store digital information in artificial DNA and recover it with perfect fidelity.
DNA storage has been attempted before. In 2012, George Church managed to convert an HTML version of a 53,400-word book he wrote into DNA code. Church’s group neatly assigned A or C to zero and G or T to one, but the problem was that repeated bases from repeated digits could introduce error (imagine trying to sequence a long string of A’s — it would be very easy to miss or add one by accident).
Goldman and Birney solved the issue of repeated bases by using a new encoding scheme. First, they converted binary digital information into ternary, which uses zero, one, and two. Then, they developed an interpretation system in which each of the digits is represented by a nucleotide base that depends on the previous one.
The scientists also broke up the DNA into fragments that can be easily manipulated. Cutting the DNA differently created overlapping sequences which provided another level of quality control. With their method fleshed out, Goldman and Birney successfully encoded and retrieved five different types of data: all of Shakespeare’s sonnets, Watson and Crick’s landmark paper, a color photograph, 26 seconds of Martin Luther King’s “I Have a Dream” speech, and the code of an algorithm used elsewhere in the study.
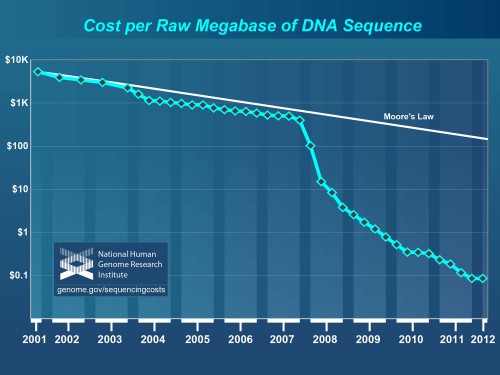
Though storing data in DNA is evidently feasible, the method has its disadvantages. The scientists spent two weeks decoding the information (though it could be done in a day with better technology). In an interview, Dr. Goldman conceded, “We’re not going to compete with silicon, I think, for speed.” He estimates that commercial implementation of their method would require about $12,400 per megabyte, which is millions of times more expensive than using magnetic tape. Yet the cost of sequencing technology is falling rapidly, and DNA storage may become economically viable soon.
For certain kinds of information, the shortcomings of DNA may be overshadowed by its benefits. The molecule stays readable after many thousands of years, provided it is kept cold, as evidenced by our sequencing of the Neanderthal genome. Furthermore, DNA is so dense that it could theoretically store all the digital information humans produce in a year in just four grams. As biological data increases exponentially, the need for compact information storage is becoming increasingly urgent; indeed, efficient data compression is already a major issue in bioinformatics. The remarkable stability and density of DNA makes it an attractive candidate for long-term information storage.

Cover Image: DNA code is composed of the nucleotide bases A, T, C and G. Courtesy of the Miraikan.
