
Researchers headed by Yale Professor of Anthropology Eric Sargis have created a family tree of mammalian species using morphological data in addition to previously used genetic data. In its morphological analysis, the study looks at physical features — especially bones — in order to find similarities between species. These similarities are then used generate a most parsimonious, or “best fit”, phylogeny of placental mammals.
The backbone of this project was the creation of MorphoBank, a web database of physical features. Researchers painstakingly coded information on 4,541 physical features for 86 mammal species, uploading both pictures and coded features. To start out, they used mostly living species because of the availability of anatomical data. However, the focus on anatomical features gives the project the potential to add data from many extinct mammal species for which genetic data cannot be obtained. Because many mammal species are extinct and can only be classified using fossils, the project will allow the fossils of extinct species to contribute to the phylogeny. The scale of this project is unprecedented and has allowed researchers to add some information to family trees that had previously been constructed on solely genetic information. Sargis describes this project by stating, “This is giving the anatomical data a chance to contribute to the answer.”
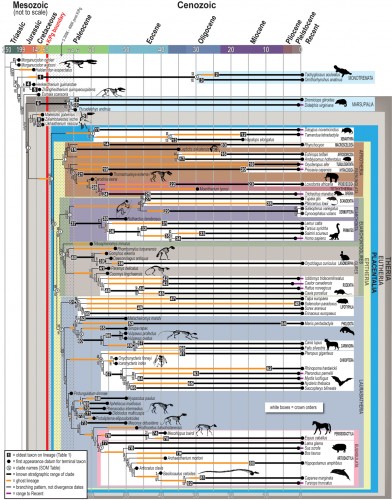
Before this paper, genetic analyses had predicted that the most recent common ancestor of placental mammals existed well before the K-Pg boundary and that over 29 mammalian lineages crossed the K-Pg boundary. This is unexpected because the K-Pg boundary refers to the Cretaceous-Paleogene extinction event — the event that knocked out the dinosaurs. It would be odd that so many mammalian lineages survived the event while many other species died. This prediction also failed to match the fossil record: As hard as scientists tried, they could not find fossils that showed the existence of diverse placental mammals before the K-Pg boundary.
With this in mind, Sargis and other researchers worked to add physical data to the phylogenetic analysis. There were originally four teams when building the matrix: one focused on the skull, one on the skeleton (not including the skull), one on soft tissue, and the other on teeth. Teeth are especially important in describing mammals, because as Sargis says, “most fossil mammals are only known from teeth”. They analyzed the 4,541 physical traits, treating each equally in the overall matrix, and came upon a number of findings that stood contrary to previous perceptions. For one thing, their analysis found that some fossil genera, like Eomaia, were probably not placental mammals. However, the main finding of the paper was that the most recent common ancestor of all placental mammals species probably arose after the K-Pg boundary and that there was a post-K-Pg increase and speciation of placental mammals.
This post K-Pg radiation has huge implications for the determination of driving factors for placental mammal speciation. Using previous dates of divergence, scientists had hypothesized that much of speciation was driven by the breakup of continents. These new data put placental radiation after the continental splits. However, even without spatial drivers, it is not that surprising that we see so many new species of placental mammals occurring after the K-Pg boundary. Sargis says, “You have lots of niches opening up. You have the extinction of dinosaurs and lots of ecological space opening up. It’s not that surprising in those ways. What is surprising is how quickly this radiation happened”.
Another interesting point from this paper was the reconstruction of the most recent common ancestor. Using all of this anatomical data, the team worked to predict what this species looked like. They found that the species was probably between 6 and 245 grams, insectivorous, and, of course, had a placenta. They were even able to predict what its teeth looked like.
Still, Professor Eric Sargis stresses, “As far as I am concerned, this is the beginning… This is a hypothesis of relationships. This is a hypothesis of the timing of divergence that will continue to be tested … as we add more taxa, especially fossil taxa, that may change the tree.” He believes that one of the most exciting parts of the project is the creation of MorphoBank, because “this will make morphological systematics much more explicit and repeatable.” This does represent somewhat of a departure from genetics-heavy techniques that are used by many labs today because of their comparative ease of use. However, this work proves that anatomical information is still just as important in determining scientific questions about the history of life.
