For many years following the earliest work on genetics, a deterministic strain of thought prevailed in biology. Conventional wisdom held that the connection between genes and traits was simple, straightforward, and above all, immutable. But in recent years, new research has muddied the waters; a picture has emerged of dynamic patterns of gene expression and complex interactions between the genome and the environment. One of the many puzzles to emerge from this updated outlook on biology is the mechanism by which some organisms make rapid changes in the genes that they express. A team of researchers at Yale University recently took on this problem and was able to determine the structure of an important chemical signal in bacteria and plants that mediates such changes. Thomas A. Steitz, the 2009 Nobel Prize Winner in Chemistry, Yuhong Zuo, and Yeming Wang used crystallographic methods to elucidate the structure of a signal molecule called guanosine pentaphosphate (ppGpp) in Escherichia coli (E. coli) bacteria. This molecule was known to be an important alarmone, a chemical that regulates transcription of DNA in response to unfavorable environmental conditions. ppGpp inhibits the synthesis of ribonucleic acid (RNA, an product of gene transcription) when nutrients are scarce and amino acids are in short supply.
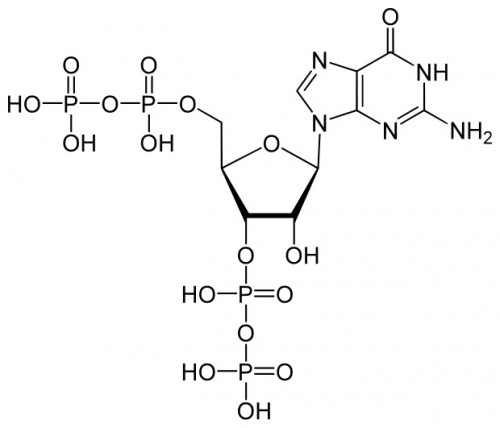
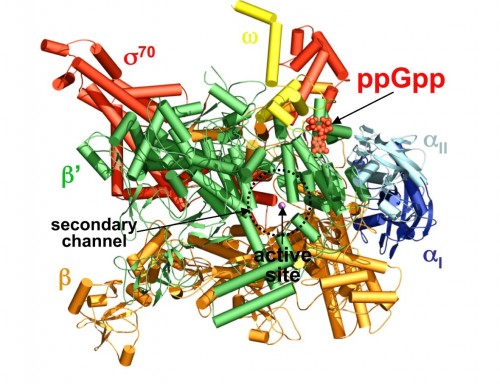
The key role played by ppGpp in allowing bacteria to adjust to harsh conditions has long made it the subject of investigation by biochemists. Professor Steitz said that while importance of ppGpp in regulating transcription was apparent and well known, his lab focused on the molecule “because it’s something else entirely to know how it does that.” The Yale team tackled that mystery by characterizing the structure of ppGpp and RNA polymerase, which synthesizes RNA, at a resolution of 4.5 angstroms (Å), a unit of measure equal to one ten-billionth of a meter. The researchers employed X-ray crystallography, a technique that uses the diffraction patterns of X-rays to measure the arrangement of atoms in a molecule, to generate the data. The crystal structures of the two molecules revealed something unexpected.According to the paper, “ppGpp bonds to a single site on the outer surface of the RNA polymerase, more than 25 Å away from the catalytic center.” This means ppGpp is unable to directly interfere with transcription. Instead, the team concluded that the alarmone blocks different modules of the RNA polymerase molecule from moving normally. In the absence of ppGpp, movement of these modules enables RNA polymerase to pull, or “ratchet,” itself along a strand of DNA. ppGpp slows or stops this ratcheting, leading to less synthesis of RNA.
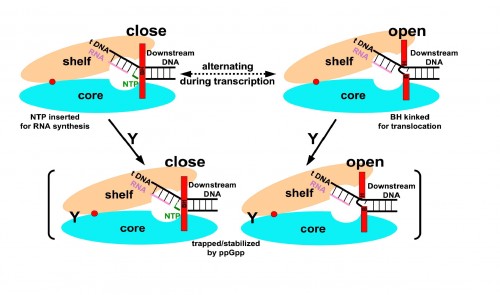
The discovery represents a significant step forward in understanding the mechanisms of ppGpp and, ultimately, the process that allows cells to “swiftly reprogram their gene expression in order to adapt to various stress conditions,” according the paper’s authors. It is also one of the first studies to show indirect regulation of transcription, accomplished by a molecule that does not block the active site of RNA polymerase. The researchers point out that transcription regulation’s ubiquity throughout nature suggests that the means by which ppGpp inhibits RNA synthesis may be common to a number of other regulatory processes. Professor Steitz explained that while knowledge of the mechanism of ppGpp has few, if any immediate applications, it may hint at a mode of regulating gene expression that is more widespread. Overall, it represents a new and intriguing facet of the complex network of biochemical interactions that brings genes to life.
Sources:
1. Yuhong Zuo, Yeming Wang, Thomas A. Steitz, The Mechanism of E. coli RNA Polymerase Regulation by ppGpp Is Suggested by the Structure of their Complex, Molecular Cell, Volume 50, Issue 3, 9 May 2013, Pages 430-436, ISSN 1097-2765, https://dx.doi.org/10.1016/j.molcel.2013.03.020.
2. Interview with Professor Thomas Steitz, interview on 10/30/2013
