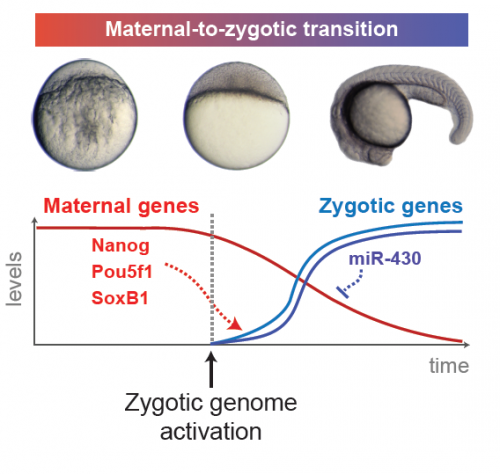
Over a nine-month period, a human baby develops from a single-celled zygote into a fully formed infant capable of seeing, crying, and eating. This carefully coordinated process requires a tightly regulated genetic program controlling the proper differentiation of each cell at every stage along the way. Although the process is initiated by maternal instructions, the newly formed zygote quickly takes over, activating its genome to begin dictating its own fate. The way by which this zygotic genome activation, a critical part of the maternal to zygotic transition, occurs is not currently well understood. However, a recent study led by Antonio Giraldez, Yale Associate Professor of Genetics, has begun to shed light on this fundamental process by identifying the necessary maternal proteins.

Development of the Embryo
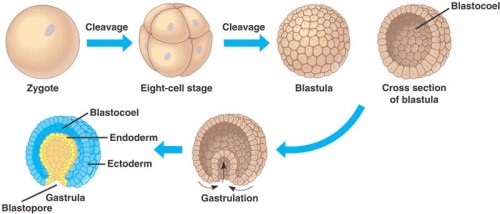
Development begins with fertilization, when a sperm enters the egg and forms the zygote. Following fertilization, cleavage occurs, in which a series of rapid cell divisions forms a bundle of small cells known as the blastula. During these divisions, the single cell multiplies into 32 cells without increasing in size; no growth occurs between each cleavage. Furthermore, up to this point, the zygotic genome is still inactive and no genes are expressed. The developing embryo is completely reliant on maternal proteins and genes to direct these early steps.
Once the blastula is formed, cell divisions change from cleavage to normal mitosis, which includes the gap phases in the cell cycle that allow the cell to grow. In order for this switch to occur, the maternal-to-zygotic transition is required. This is “a transition point when the maternal instructions are no longer enough for development, when the embryo starts to follow some of its own instructions,” said post-doctoral researcher Miler Lee, co-first author of the paper. The shift involves activation of the zygotic genome and clearance of maternal products, marking a turning point in the independence of the embryo. While many of the mechanisms involved have been studied, the specific maternal proteins responsible for initiating the transition are not yet known.
Identifying these protein factors was the goal of the Giraldez Lab’s study. The study used zebrafish, a model organism favored in developmental biology for its transparent embryos which develop outside the body. The research team began by sequencing all of the RNA within the embryo to determine which zygotic genes were the first to be transcribed, or activated. By pinpointing these genes, the researchers defined potential targets for maternal factors involved in zygotic genome activation. Graduate student and co-first author Ashley Bonneau explained that, because maternal messenger RNA (mRNA) should be fully processed, or spliced, into its mature form, if “we look for sequences that should already be processed out, we can hypothesize that transcripts with these sequences are the zygotic transcripts.”
This analysis resulted in the identification of over 5000 genes which had been transcribed within four hours after fertilization. To narrow this field down to genes whose expression is actually activated by maternal as opposed to zygotic factors, the scientists specifically blocked splicing of zygotic mRNA, thereby preventing their translation into proteins. When only the maternal factors were able to act, transcription of 269 genes was still detected, representing the “first wave” of zygotic gene expression.

Activating the First Wave
Once the team had identified the genes activated by maternal products, the question became which products were responsible for their activation. To this end, ribosome profiling, which sequences mRNAs bound to cellular units responsible for producing proteins, was used to determine which maternal mRNAs for transcription factors were being translated most frequently into proteins. Transcription factors, proteins which regulate the expression of other genes, are likely to be important players in activating the zygotic genome.
The profiling identified three frequently translated transcription factors: Nanog, Sox19b, and Pou5f1. 86 percent of zygotic genes showed reduced expression levels when the researchers blocked the production of these factors. Even more significantly, more than 95 percent of treated embryos experienced complete developmental arrest. However, although these factors are crucial, they do not account for all gene activation according to the team’s analysis. Bonneau believes that the remaining 14 percent may not have shown reduced activation due to a combination of reasons. “It may be that there was only a partial loss of function [of Nanog, Sox19b, and Pou5f1], that other transcription factors are also essential for this process to occur, and that the landscape that this event is taking place in is important. It’s not just that the factors have to be present, but the chromatin — how the DNA is actually packaged — has to be in a proper state for the activation to occur,” she said.
One gene that was markedly reduced in the absence of Nanog, Sox19b, and Pou5f1 was the microRNA (miRNA) miR-430. miRNAs are small, noncoding RNAs that function to silence other gene products. miR-430 in particular binds to many maternal RNAs in the zygote, resulting in their degradation and the clearance of the maternal genetic program. “Maternal instructions in the form of mRNAs are very important for the early stages of embryogenesis, but as [the embryo] transitions towards differentiation, a lot of those RNAs become unnecessary,” said Lee. “miR-430 hastens this process of getting rid of these unnecessary products […] so that cellular resources are focused more on things that are necessary moving forward.” Therefore, regulating miR-430 expression is a key part of Nanog, Sox19b, and Pou5f1’s roles in setting embryonic development in motion.
Cleaning the Slate
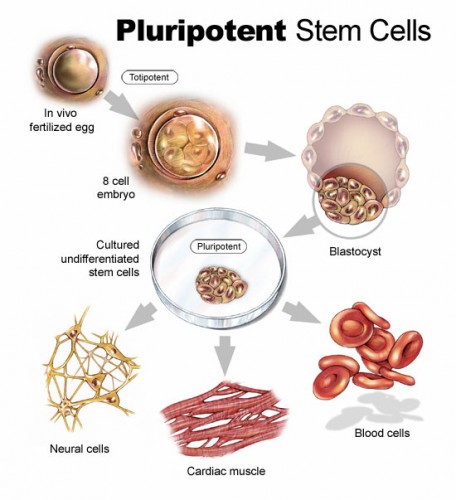
Interestingly, the same factors Nanog, Sox19b, and Pou5f1 have been identified by previous studies as the main regulators of pluripotency, or the ability to differentiate into many different cell types, in stem cells. This link may provide deeper insight into what is occurring during the maternal-to-zygotic transition. According to Bonneau, “With iPS (induced pluripotent stem] cells, you’re taking a differentiated cell, clearing away [its] instructions […] and reprogramming it into an iPS cell that has a different transcriptional landscape. That’s what we think is occurring at [the transition] — it’s a reprogramming of the embryo from one state to another to allow development to proceed.” The connection between embryonic development and stem cell pluripotency suggests that the three factors may represent a fundamental pathway to “clean the slate” of a cell and allow it to take on a new genetic program.
In the future, Lee and Bonneau plan to investigate the role of Nanog, Sox19b, and Pou5f1 on a molecular level to see how they interact with other proteins to activate the first wave of zygotic transcription. They are also examining the timing of the recruitment of the factors to determine if one of them needs to arrive before the others to facilitate an interaction. By completing these further studies, they hope to gain a more detailed comprehension of how Nanog, Sox19b, and Pou5f1 orchestrate the first steps in the development of life.
About the Author:
Grace Cao is a sophomore Molecular, Cellular and Developmental Biology major in Timothy Dwight College. She is a copy editor for the Yale Scientific Magazine and works in Professor Carla Rothlin’s lab in the Immunobiology Department.
Acknowledgements:
The author would like to thank Antonio Giraldez, Miler Lee, and Ashley Bonneau for their time and clear explanations of their research on the maternal to zygotic transition.
