Evolution is the process by which all life on Earth has differentiated itself. It is what has separated whales from willows and butterflies from bacteria. Mutation drives this process by introducing occasional mistakes in gene sequences. These errors sometimes give an organism a selective advantage, and are thus passed down to later generations through natural selection. Many of us are familiar with Darwin’s birds and the way different beaks types are suited to specific food sources. Perhaps less familiar is the idea that evolution happens at every level in the hierarchy of life — from plants and animals right down to the molecules that make up an organism.
Researchers have extensively analyzed the mutations of individual genes. They have mapped how these genetic hiccups enhance or weaken the fitness of an organism. But does all mutation arise at the gene level? Or could evolution also be influenced by mutations in what surrounds a gene, by mutations in gene networks? Our understanding of this process is more limited.
A gene regulatory network is a system of DNA strands and proteins that interact to control gene expression in an organism. Gene networks evolve through mutations in the same way that any other component of an organism evolves. Despite this, the specific mechanism of evolution in gene regulatory networks was unknown to scientists. That is, until a team of Yale researchers published findings that suggest an answer. Murat Acar, an assistant professor of molecular, cellular, and developmental biology and physics, and postdoctoral associates Weilin Peng, Ping Liu, and Yuan Xue explored potential mechanisms for the evolution of gene networks. They concluded that promoters and their negative feedback loops are key. Moreover, this research expands our knowledge of evolution as it occurs at every level of life.

The team investigated regulatory elements that lie close to genes as part of the gene network. These cis-regulatory elements are segments of DNA that do not code for proteins, but that instead regulate the expression of genes downstream. The genetic regions that directly precede these genes are called promoters. Acar’s finding on the significant role that promoters play in evolutionary processes sheds light on the fact that individual gene mutations do not provide a complete picture. His study opens the door for further explorations into mutations of cis-regulatory elements that factor into evolution.
Acar’s research is an exciting story of genetic discovery, one that ends with a promoter called GAL80 and its negative feedback loop. Of course we must start at the beginning — with two species of yeast and a sugar, galactose.
The galactose utilization network
The Yale team decided to focus on the galactose utilization network. Also called the GAL network, it is responsible for metabolizing the simple sugar galactose. Two species of yeast were used in the experiments: Saccharomyces cerevisiae and Saccharomyces paradoxus. The species are highly similar, both containing GAL networks with comparable structures. These GAL networks contain four regulatory promoters — GAL2, GAL3, GAL4, and GAL80 — each controlling expression of its respective protein. Since these proteins serve the same functional roles in both species, the researchers were able to replace the network components in S. cerevisiae with their genetic counterparts from S. paradoxus. A promoter from the first species could be inserted into the genome of the second, and vice versa. By swapping promoters, the scientists were probing one critical question: Is it only individual gene mutations that cause evolutionary change, or can mutated elements of a gene network have the same effect?
The galactose network was an ideal subject of study because it is already well-characterized. The experimenters wanted to start out with as few unknowns as possible, given that the answers they were after were so mysterious. The question was how differences between genetic counterparts affect the gene network activity in each species. “Whenever your goal is to understand evolution at the gene network level, it is essential to have a model gene network with well-characterized components,” Acar said. The choice of the GAL network, then, was a control in this experiment — one less variable to distract from the manipulated variable of swapped promoters.
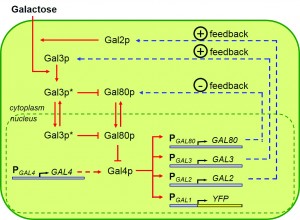
Image courtesy of Murat Acar.
Negative feedback
A negative feedback loop occurs when some product of a pathway circles back to inhibit an initial step, thereby shutting down the pathway or mildly reversing its effect. For example, our body temperature is maintained via negative feedback: We shiver to warm up when we are cold, and we sweat to cool down if the temperature is too high. Of the four regulatory promoters in the GAL network, GAL80 is the only one that mediates a negative feedback loop. It dictates whether GAL network activity will be on or off.
GAL80 proteins act as repressors in the galactose network, while GAL4 proteins are activators. Having a large amount of GAL80 proteins switches off gene network activity by deactivating GAL4, while having few GAL80 proteins present allows GAL4 to function unrepressed, sustaining network activity.
Acar and his colleagues manipulated these regulatory elements in their yeast by swapping promoters between the two species. They replaced one, two, three, or all four of the promoters, yielding 16 combinations in total. Three of the regulatory promoters — GAL2, GAL3, and GAL4 — control other functions, such as positive feedback loops, and none of these elicited much of a change in network activity when replaced. In contrast, replacing the GAL80 promoter alone caused a dramatic change in network activity, which suggests that GAL80 and its associated negative feedback pathway are both players in a mechanistic explanation of gene network evolution.
The key to understanding how gene network activity can evolve lies in a certain motif. Now, after the results from Acar’s study, this motif appears to be the negative feedback loop.
How to measure differences in gene network activity is a complicated question. The negative feedback loop, mediated by the GAL80 promoter, has a strong influence on the yeast cell’s ability to flip between on and off states. “On” cells are able to metabolize galactose, while “off” cells cannot. In a solution with an extremely high concentration of galactose, almost every cell will be on. Conversely, in a solution with no galactose, every cell will be off.
The ratio of on cells to the total number of cells in a population of yeast describes its inducibility, which was an important data point for Acar’s team. It also provides a phenotype — a visible expression of genes, such as brown eyes or curly hair — that allows researchers to observe changes in gene network activity. For the yeast species at hand, the phenotype was the fraction of cells metabolizing galactose, highlighted by a special type of indicator called the yellow fluorescent protein. Through these methods, the researchers could use levels of galactose and of galactose metabolism as proxies for the gene network’s activity.
Image courtesy of the University of California San Diego, Julian Schroeder Lab.
Plotting the galactose concentration of the yeasts’ environment against the inducibility yields what is called an inducibility profile. Despite the similarity in GAL network structures between species, it turned out that the yeast showed significantly different inducibility profiles, meaning there must be some cross-species difference in components of the GAL network. But how are they different? The researchers considered two possibilities: either the proteins themselves behave differently, or the number of proteins is different between the two types of yeast. It was a question of quality versus quantity — perhaps proteins can more effectively transmit the galactose signal in one species due to enhanced protein activity, or maybe there are simply more of these proteins in one species compared to the other.
To find out whether quantity or quality was responsible for the observed differences in gene network activity, Acar’s team had to decide which parts of the network to study. Promoters control the abundance of proteins, while specific genes control the characteristics of proteins. The researchers chose to focus on the former. “We already knew that the promoters were doing a lot of very novel, interesting things,” Acar said, referring to some of his previous research on how promoters can control various characteristics of the GAL network. With this in mind, Acar’s group hypothesized that promoters might also play a role in how the networks of these two yeast species diverged through evolution.
Understanding evolution, and more
The results from this research not only help to decipher the codes of genetic evolution, but are also useful in understanding antibiotic resistance. If a mistake in an organism’s genome ends up helping it battle against a poison, that mutation can be passed down to successive generations. These later generations then carry the gene that offered resistance to the poison in the first place. This is how so many disease-causing organisms develop resistance to antibiotics. While it is important to understand how individual genes may contribute to antibiotic resistance, these alone do not tell the whole story. A gene’s expression is inextricably influenced by the network in which it is embedded. Understanding how entire gene networks evolve to resist antibiotics may inform more effective strategies to prevent the development of resistant strains of diseases.
Acar’s research is part of a small wave of new studies that provide empirical evidence on gene network evolution. Both the quantitative nature of the study and the fact that it draws a connection between gene network activity and promoters that control feedback make it an important contribution to understanding an evolutionary mechanism. Of course, this paper is only the beginning. “Time will show us whether or not this is unique to the GAL network or one of those design principles that evolution operates at the gene network level,” Acar said.
The powerful potential of a negative feedback loop has far-reaching implications. Negative feedback loops are everywhere in gene networks — not just in yeast, and not just the GAL network. They can be found in the gene networks of oak trees, bulldogs, and humans. With more tests on different gene networks in different organisms, perhaps a pattern will emerge that links negative feedback to the evolution of gene networks on the whole. This ongoing work will hopefully provide us with a better understanding of how mechanisms of genetic evolution themselves evolve.
About the Author: Genevieve Sertic is a freshman prospective electrical engineering major in Pierson College. She is a Copy Editor for the Yale Scientific Magazine and works with Yale’s solar energy student group, Project Bright, to promote solar power on campus.
Further Reading: Acar, Murat, Bernardo Pando, Frances Arnold, Michael Elowitz, and Alexander Van Oudenaarden. 2010. “A General Mechanism for Network-Dosage Compensation in Gene Networks.” Science 329 (5999): 1656–1660. doi: 10.1126/science.1190544
Cover Image: Art by Stephanie Mao.
Acknowledgements: The author would like to thank Dr. Murat Acar for his time and enthusiasm for his research.