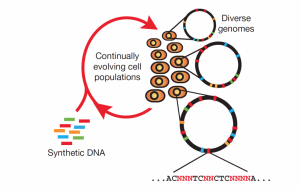
Cracking and manipulating the genetic code has been the holy grail of geneticists for decades. Recent advancements in genome-wide editing technology have led to a new wave of genetically engineered organisms that have their entire genomes recoded, rather than just one or two genes. Working towards this end is a team led by Assistant Professor of Molecular, Cellular, and Developmental Biology Farren Isaacs. At their research facility on Yale University’s West Campus, the team has used multiplex automated genome engineering (MAGE) to alter the genomic code of Escherichia coli bacteria so that they are more resistant to viral infections.
Since viruses are unable to reproduce by themselves, they typically infect a host cell and hijack a host’s cellular machinery in order to reproduce. In bacteria, this process is expedited when viruses are propagated through horizontal gene transfer (HGT), a natural process that allows bacteria to exchange genetic material. HGT increases the overall genetic diversity of a bacterial population but also makes it susceptible to widespread viral infections. This can be especially problematic for biofactories, production plants that use enormous vats of bacteria to mass-produce pharmaceutical products. Viral contamination in biofactories rapidly spreads through bacterial populations and is a costly recurring problem. In 2009, a viral contamination incident at a Genzyme plant in Massachusetts shut the facility down for weeks, causing supply constraints of important drugs and up to $300 million in losses.
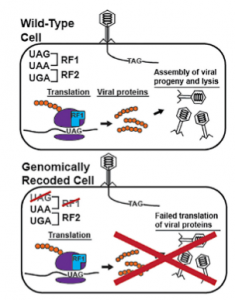
In their Cell Systems paper published in August 2016, Isaacs and Natalie Ma, a doctoral student at Yale University, demonstrated how a genomically recoded strain of E. coli lacking UAG stop codons exhibited reduced HGT activity. The viability of the recoded E. coli strain was unaffected since all the UAG stop codons were replaced with functionally equivalent UAA stop codons. The recoding, however, did impart resistance to multiple viruses. “Because the genetic code of the virus is now incompatible with the changed code of [the] host, the virus then is not able to faithfully express all of its genes,” Isaacs said. “This obstructs the ability of the virus to mass produce itself, release its progeny into the environment, and repeat the process,” he added.
Despite the recoded E. coli’s unexpectedly strong viral resistance, it was not completely immune. Over time, due to the high mutation rate of viruses and selective pressure from the recoded E. coli, the viruses were able to adapt to the lack of UAG stop codons and regain the ability to infect the genomically recoded bacteria. As Ma and Isaacs have only recoded a single stop codon in these bacteria, they anticipate that further recoding of the E. coli’s genome would make it nearly impossible for the viruses to adapt to the new genomic code.
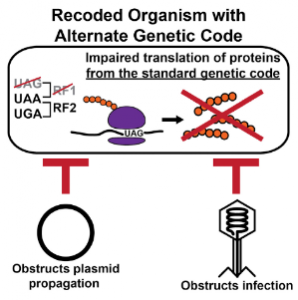
Using similar genomically recoded virus-resistant bacteria in biofactories would make drug production much more cost-effective and easier to manage. In addition, because such recoded microbial communities are genetically isolated, genome recoding could also function as a method of stabilizing GMO populations and safely deploying them into open systems, mitigating concerns about GMOs breeding with the wild-type population. In order to be impactful, this research must transcend the lab bench. “Society needs to help figure out what is safe and what level of risk is acceptable,” Ma said. In reckoning with genetically modified organisms, scientists and the public will have to work together.

