
Thanks to the power of computer programming, we can control many actions with extreme precision, from the scenery of a video game to the movements of a surgical apparatus. These actions are all defined by a code with a set of precise rules — a code quite similar in function to biology’s own DNA. Now, biochemists at the University of Washington have begun harnessing the coding power of DNA to precisely control chemical reactions, just as computer code controls the virtual world of a video game.
Like computer programs that direct machines to undertake specific and repeatable tasks, DNA instructs cellular machinery to undertake a specific set of chemical processes. However, a fundamental difference between computer programs and biochemical systems is that the latter are much more difficult for scientists to control. While modifying a computer program often involves only minor adjustments, modifying a biochemical system currently requires rebuilding that system from scratch.
For decades, scientists have realized the potential of “programming” chemical reactions, but the difficulty of precisely controlling molecules has hindered any major breakthroughs until now. This year, a team of engineers from the University of Washington, led by Assistant Professor of Engineering Georg Seelig, set out to create a system for studying and fine-tuning chemical reactions with DNA — a programmable biological computer of sorts.
“I think this [approach] is appealing because it allows you to solve more than one problem,” said Seelig in a September University of Washington Today article. “If you want a computer to do something else, you just reprogram it. This project is very similar in that we can tell chemistry what to do.”
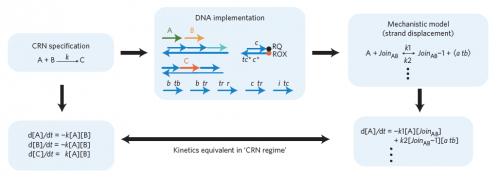
The basis of this DNA programming was simple, but its execution proved to be more complex. The “instructions” the researchers provided to the system were custom-designed strands of DNA. These DNA strands were allowed to react with one another and form new combinations, just as a computer processes the programmer’s instructions to generate output. Since the four subunits of DNA follow a basic base-pairing rule — adenine binds to thymine, and cytosine binds to guanine — the team could precisely control the outcome of the reaction by simply tweaking their original input. This can involve changing the sequences of certain DNA strands, or adjusting how much of each strand gets mixed into the “computational soup.”
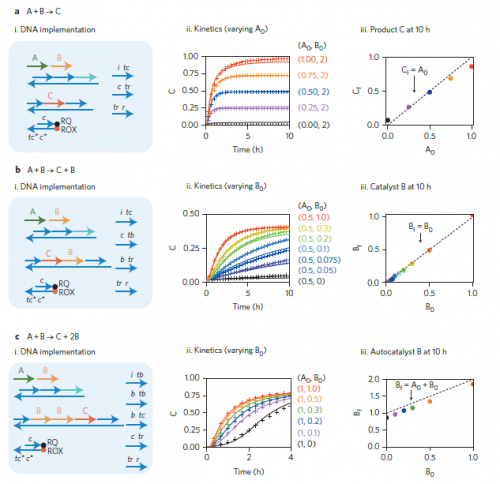
The reactions that Seelig’s team programmed used the basic language of chemistry. For example, A + B → C + D meant that signals A and B should be transformed to signals C and D. The researchers proved that this “program” produced C and D not only reliably but also in the right amounts and at the predicted rate. Thus, these programmed chemical reactions behaved exactly as they would in a native environment. In addition to controlling simple, one-step processes, the team also programmed entire reaction cascades, in which the output of one reaction becomes the input for the next. Again, these reactions produced the expected output at the predicted rate and quantity.
Seelig’s team then used their DNA reactions to mimic the function of a real computer: by applying an algorithm, the DNA program was able to solve a complex computing problem. Specifically, it could compare the concentrations of two inputs, X and Y, and decide which one was in the majority. While this computing problem seems simple for a human brain to decipher, engineered DNA strands that are capable of making decisions represent a step forward in the field of molecular programming.
“We start from an abstract, mathematical description of a chemical system and then use DNA to build the molecules that realize the desired dynamics,” said Seelig. “The vision is that eventually, you can use this technology to build general-purpose tools.”
As the field of DNA programming is still in its early stages, this new approach is not yet ready for application to the medical field. However, researchers hope that learning to program more complex chemical reaction systems will have important applications. For example, this molecular programming technique could be used to make molecules that self-assemble within living cells. These could act as “smart” sensors that detect specific abnormalities and respond accordingly, perhaps by direct drug delivery to the abnormal cells.
Seelig’s team recently received $2 million from the National Science Foundation as part of an initiative to spur research in molecular programming. Seelig and his team will use this funding to continue their work towards applications that may change the face of biotechnology.
Cover Image: An engineering team at the University of Washington aims to “program” chemical reactions by controlling DNA. Courtesy of Trunews.
